Indice
HPC.unipr.it User Guide
Login
In order to access the resources, you must be included in the LDAP database of the HPC management server.
Any user needs to be associatated to a research group. The current list of research groups is available here.
Requests for new access must be sent to sti.calcolo<at>unipr.it by the contact person of the research group.
Any request must be validated by the representative in the scientific committee for the research area.
Once enabled, the login is done through SSH on the login host:
ssh name.surname@login.hpc.unipr.it
Password access is allowed only within the University network (160.78.0.0/16). Outside this context it is necessary to use the University VPN.
Furthermore we suggest to use public key authentication.
Password-less access among nodes
In order to use the cluster it is necessary to eliminate the need to use the password among nodes, using public key authentication. It is necessary to generate on login.hpc.unipr.it the pair of keys, without passphrase, and add the public key in the authorization file (authorized_keys):
Key generation. Accept the defaults by pressing enter:
ssh-keygen -t rsa -P '' -f ~/.ssh/id_rsa
Copy of the public key into authorized_keys:
cat ~/.ssh/id_rsa.pub >> ~/.ssh/authorized_keys
Change the authorized_keys file permission:
chmod 0600 ~/.ssh/authorized_keys
Access with public key Authentication
The key pair must be generated with the SSH client. The private key should be protected by an appropriate passphrase (it is not mandatory but recommended). The public key must be included in your authorized_keys file on the login host.
An authorized_keys file contains a list of OpenSSH public keys, one per line. There can be no linebreaks in the middle of a key, and the only acceptable key format is OpenSSH public key format, which looks like this:
ssh-rsa AAAAB3N[... long string of characters ...]UH0= key-comment
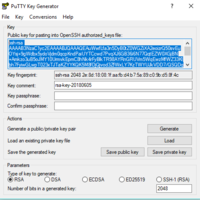
If you use the SSH client for Windows PuTTY (http://www.putty.org), you need to generate the public and private key pair with PuTTYgen and save them into 2 separate files.
The private key, with extension .ppk, must be included in the Putty (or WinSCP) configuration panel:
Configuration -> Connection -> SSH -> Auth -> Private key file for authentication
Once done on Putty, return to Session:
Session -> Host name: <name.surname>@login.hpc.unipr.it -> Copy and Paste into "Saved Session" -> Save -> Open
The public key can be copied from the PuttyGen window (see picture below) and included in the .ssh/authorized_keys file on login.hpc.unipr.it:
cd .ssh nano authorized_keys ctrl + X - confirm and press Enter
File transfer
SSH is the only protocol for external communication and can also be used for file transfer.
If you use a Unix-like client (Linux, MacOS X) you can use the command scp or sftp.
On Windows systems, the most used tool is WinSCP (https://winscp.net/eng/docs/introduction). During the installation of WinSCP it is possible to import Putty profiles.
A good alternative is SSHFS.
Graphical User Interface
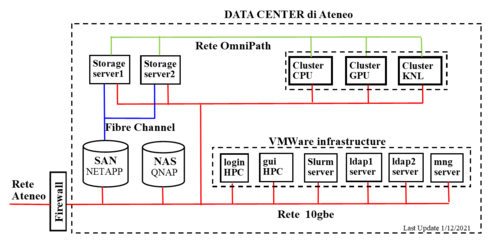
Hardware
Cluster architecture:
Computing nodes
| Partition | Node Name | CPU Type | HT | #Cores | MEM (GB) | GPU | Total | Owner |
|---|---|---|---|---|---|---|---|---|
| cpu | wn01-wn08 | 2 Intel Xeon E5-2683v4 2.1GHz 16c | NO | 32 | 128 | 0 | 256 cores | Public |
| cpu | wn11-wn14, wn17 | 2 Intel Xeon E5-2680v4 2.4GHz 14c | NO | 28 | 128 | 0 | 140 cores | Public |
| cpu | wn18-wn19 | 4 Intel Xeon E5-6140 2.3GHz 18c | NO | 72 | 384 | 0 | 144 cores | Public |
| cpu | wn33 | 2 Intel Xeon E5-2683v4 2.1GHz 16c | NO | 32 | 1024 | 0 | 32 cores | Public |
| cpu | wn34 | 4 Intel Xeon E7-8880v4 2.2GHz 22c | NO | 88 | 1024 | 0 | 88 cores | Public |
| cpu | wn35-wn36 | 4 Intel Xeon E5-6252n 2.3 GHz 24c | NO | 96 | 512 | 0 | 192 cores | Public |
| cpu | wn80-wn95 | 2 AMD EPYC 7282 2.8 GHz 16c | NO | 32 | 256 | 0 | 512 cores | Public |
| cpu | wn96-wn98 | 2 AMD EPYC 7282 2.8 GHz 16c | NO | 32 | 512 | 0 | 96 cores | Public |
| cpu_mm1 | wn20 | 4 Intel Xeon E5-6140 2.3GHz 18c | NO | 72 | 384 | 0 | 72 cores | Private |
| cpu_mm1 | wn27-wn30 | 4 AMD EPYC 7413 2.65GHz 24c | NO | 24 | 192 | 0 | 72 cores | Private |
| cpu_infn | wn21 | 4 Intel Xeon E5-6140 2.3GHz 18c | NO | 72 | 384 | 0 | 72 cores | Private |
| cpu_infn | wn22 | 4 Intel Xeon E5-5218 2.3 GHz 16c | NO | 64 | 384 | 0 | 64 cores | Private |
| cpu_bioscienze | wn23 | 4 Intel Xeon E5-5218 2.3 GHz 16c | NO | 64 | 384 | 0 | 64 cores | Private |
| cpu_mmm | wn24-wn25 | 4 Intel Xeon E5-5218 2.3 GHz 16c | NO | 64 | 384 | 0 | 128 cores | Private |
| cpu_dsg | wn26 | 2 Intel Xeon Silver 4316 CPU 2.3 GHz 20c | NO | 40 | 512 | 0 | 40 cores | Private |
| cpu_guest | wn20-wn21 | 4 Intel Xeon E5-6140 2.3 GHz 18c | NO | 72 | 384 | 0 | 144 cores | Guest |
| cpu_guest | wn22-wn25 | 4 Intel Xeon E5-5218 2.3 GHz 16c | NO | 64 | 384 | 0 | 256 cores | Guest |
| cpu_guest | wn27-wn30 | 4 AMD EPYC 7413 2.65GHz 24c | NO | 24 | 192 | 0 | 256 cores | Guest |
| gpu | wn41-wn42 | 2 Intel Xeon E5-2683v4 2.1 GHz 16c | NO | 32 | 256 | 6 P100 | 12 GPU | Public |
| gpu | wn44 | 2 AMD EPYC 7352 2.3 GHz 24c | NO | 48 | 512 | 4 A100 40G | 4 GPU | Public |
| gpu | wn45 | 2 AMD EPYC 7352 2.3 GHz 24c | NO | 48 | 512 | 8 A100 80G | 8 GPU | Public |
| gpu_hylab | wn44 | 2 AMD EPYC 7352 2.3 GHz 24c | NO | 48 | 512 | 2 V100, 2 A100 80G | 4 GPU | Private |
| gpu_vbd | wn46 | 2 AMD EPYC 7313 3.0 GHz 16c | NO | 32 | 512 | 3 A100 80G, 1 Quadro RTX 6000 | 4 GPU | Private |
| gpu_vbd | wn49 | 2 AMD EPYC 9354 3.25 GHz 32c | NO | 32 | 768 | 8 L40S 48G | 4 GPU | Private |
| gpu_ibislab | wn47 | 2 AMD EPYC 7413 2.65 GHz 24c | NO | 48 | 512 | 8 A30 24G | 8 GPU | Private |
| gpu_mm1 | wn48 | 2 AMD EPYC 7413 2.65 GHz 24c | NO | 48 | 512 | 8 RTX A5000 24G | 8 GPU | Private |
| gpu_fisstat | wn50 | 2 Intel Xeon Gold 6426Y 16c | NO | 32 | 512 | 4 L40S 48G | ||
| gpu_guest | wn44 | 2 AMD EPYC 7352 2.3 GHz 24c | NO | 48 | 512 | 2 V100, 2 A100 80G | 4 GPU | Guest |
| gpu_guest | wn46 | 2 AMD EPYC 7313 3.0 GHz 16c | NO | 32 | 512 | 3 A100 80G, 1 Quadro RTX 6000 | 4 GPU | Guest |
| gpu_guest | wn47 | 2 AMD EPYC 7413 2.65 GHz 24c | NO | 48 | 512 | 8 A30 24G | 8 GPU | Guest |
| gpu_guest | wn48 | 2 AMD EPYC 7413 2.65 GHz 24c | NO | 48 | 512 | 8 RTX A5000 24G | 8 GPU | Guest |
| vrt | wn61-wn64 | 2 Intel Xeon Gold 6238R 2.2GHz 28c | YES | 8 | 64 | 0 | 32 cores | Public |
[I nodi privati sono a disposizione dei proprietari attraverso code private (cpu_mm1, cpu_infn, gpu_hylab). Se i nodi sono liberi gli altri utenti possono accedere utilizzando code "guest" (cpu_guest, gpu_guest). I job in esecuzione nelle code guest vengono interrotti e risottomessi se arriva un job nella coda privata.]
Owner can use their nodes using private queues/partitions (cpu_mm1, cpu_infn, gpu_hylab). If these nodes are free, other users can use them using "guest" partition (cpu_guest, gpu_guest). But, if a owner's job arrive, running guest jobs are stopped and requeued.
GPUs
| Node Name | GPU Type |
|---|---|
| wn41-wn42 | NVIDIA Corporation GP100GL [Tesla P100 PCIe 12GB] (rev a1) |
| wn44 | NVIDIA Corporation GV100GL [Tesla V100 PCIe 32GB] (rev a1) |
| wn44 | NVIDIA Corporation GA100 [Grid A100 PCle 40GB] (rev a1) |
| wn44 | NVIDIA Corporation Device 20b5 [A100 PCIe 80GB] (rev a1) |
| wn45 | NVIDIA Corporation Device 20b5 [A100 PCIe 80GB] (rev a1) |
| wn46 | NVIDIA Corporation Device 20b5 [A100 PCIe 80GB] (rev a1) |
| wn46 | NVIDIA Corporation TU102GL [Quadro RTX 6000/8000] (rev a1) |
| wn47 | NVIDIA Corporation Device 20b7 [A30 PCIe 24GB] (rev a1) |
| wn48 | NVIDIA Corporation Device 2231 [RTX A5000 24GB] (rev a1) |
| wn49-wn50 | NVIDIA Corporation Device 26b9 [L40S 48GB] (rev a1) |
| wn51 | NVIDIA Corporation Device 2321 [H100 NVL 96GB] (rev a1) |
Peak Performance
| Code Nade | Device | TFlops (d.p) | notes |
|---|---|---|---|
| Broadwell | 2 Intel Xeon E5-2683v4 2.1GHz 16c | 0.54 | 2 x 16 (cores) x 2.1 (GHz) x 2 (add+mul) x 4 (256 bits AVX2 / 64 bits d.p.) |
| 2 Intel Xeon E5-2680v4 2.4GHz 14c | 0.54 | 2 x 14 (cores) x 2.4 (GHz) x 2 (add+mul) x 4 (256 bits AVX2 / 64 bits d.p.) | |
| 4 Intel Xeon E7-8880v4 2.2GHz 22c | 1.55 | 4 x 22 (cores) x 2.2 (GHz) x 2 (add+mul) x 4 (256 bits AVX2 / 64 bits d.p.) | |
| Skylake | 4 Intel Xeon E5-6140 2.3GHz 18c | 2.65 | 4 x 18 (cores) x 2.3 (GHz) x 2 (add+mul) x 8 (512 bits AVX2 / 64 bits d.p.) |
| Cascade Lake | 4 Intel Xeon E5-6252n 2.3 GHz 24c | 3.53 | 4 x 24 (cores) x 2.3 (GHz) x 2 (add+mul) x 8 (512 bits AVX2 / 64 bits d.p.) |
| 4 Intel Xeon E5-5218 2.3 GHz 16c | 2.36 | 4 x 16 (cores) x 2.3 (GHz) x 2 (add+mul) x 8 (512 bits AVX2 / 64 bits d.p.) | |
| 2 Intel Xeon Gold 6238R 2.2GHz 28c | 0.56 | 2 x8 (cores) x 2.2 (GHz) x 2 (add+mul) x 8 (512 bits AVX2 / 64 bits d.p.) | |
| Ice Lake | 2 Intel Xeon Silver 4316 CPU 2.3 GHz 20c | 1.47 | 2 x 20 (cores) x 2.3 (GHz) x 2 (add+mul) x 8 (512 bits AVX2 / 64 bits d.p.) |
| Rome | 2 AMD EPYC 7282 2.8 GHz 16c | 1.43 | 2 x 16 (cores) x 2.8 (GHz) x 2 (add+mul) x 8 (2x256 bits AVX2 / 64 bits d.p.) |
| Milan | 1 AMD EPYC 7413 2.65 GHz 24c | 1.02 | 1 x 24 (cores) x 2.65 (GHz) x 2 (add+mul) x 8 (2x256 bits AVX2 / 64 bits d.p.) |
| 1 GPU Quadro RTX 6000 | 0.51 | 16.3 TFlops single, 32.6 TFlops half/tensor | |
| 1 GPU P100 | 4.70 | 9.3 TFlops single, 18.7 TFlops half/tensor | |
| 1 GPU V100 | 7.00 | 14 TFlops single, 112 TFlops half/tensor | |
| 1 GPU A100 | 9.70 | 19.5 TFlops single, 312 TFlops half/tensor | |
| 1 GPU A30 | 5.20 | 10.3 TFlops single, 165 TFlops half/tensor | |
| 1 GPU RXT A5000 | 0.42 | 27.8 TFlops single, 222.2 TFlops half/tensor | |
| 1 GPUL40S | N/A | 91.6 TFlops single, 366 TFlops half/tensor |
Nodes Usage (intranet: authorized users only)
Node Interconnection Intel OmniPath
Peak performance:
Bandwidth: 100 Gb/s, Latency: 100 ns.
Software
The operating system for all types of nodes is CentOS 7.X.
Environment Software (libraries, compilers e tools): Listglobale o List
Some software components must be loaded in order to be used.
To list the available modules:
module avail
To upload / download a module (example intel):
module load intel module unload intel
To list the loaded modules:
module list
To unload all the loaded module:
module purge
Storage
The login node and computing nodes share the following storage areas:
| Mount Point | Env. Var. | Backup | Quota | Usage | Technical details |
|---|---|---|---|---|---|
| /hpc/home | HOME | yes | 50 GB | Programs and data | GPFS SAN near line |
| /hpc/group | GROUP | yes | 100 GB | Programs and data | GPFS SAN near line |
| /hpc/scratch | SCRATCH | no | max 1 month | run-time data | GPFS SAN |
| /hpc/archive | ARCHIVE | yes | 400 GB | Archive | GPFS SAN near line |
| /hpc/share | Application software and database | SAN near line |
Checking disk usage in a dir and sub-dir:
du -h
Checking personal disk quota and usage on /hpc/home:
hpc-quota-user
Checking disk quota and usage of the group disk on /hpc/group:
hpc-quota-group
Checking disk quota and usage of the archive disk on /hpc/archive:
hpc-quota-archive
Acknowledgement
This research benefits from the HPC (High Performance Computing) facility of the University of Parma, Italy
The authors are requested to communicate the references of the publications, which will be listed on the site.
Job Submission with Slurm
The queues are scheduled with Slurm Workload Manager.
Slurm Partitions
| Partition | QOS | Nodes | Resources | TimeLimit | Max Running | Max Submit | Max nodes | Audience |
|---|---|---|---|---|---|---|---|---|
| per user | per user | per job | ||||||
| cpu | cpu | wn[01-19,33-36,80-98] | 1516 core | 3-00:00:00 | 24 | 2000 | 8 | Public |
| cpu_mm1 | cpu_mm1 | wn20 | 72 core | 3-00:00:00 | 36 | 2000 | 1 | Private |
| cpu_infn | cpu_infn | wn[21-22] | 136 core | 3-00:00:00 | 36 | 2000 | 2 | Private |
| cpu_bioscienze | cpu_bioscienze | wn23 | 64 core | 3-00:00:00 | 36 | 2000 | 1 | Private |
| cpu_mmm | cpu_mmm | wn[24-25] | 128 core | 7-00:00:00 | 36 | 2000 | 2 | Private |
| cpu_dsg | cpu_dsg | wn26 | 40 core | 7-00:00:00 | 36 | 2000 | 1 | Private |
| cpu_guest | cpu_guest | wn[20-26] | 440 core | 1-00:00:00 | 24 | 200 | 4 | Guest |
| gpu | gpu | wn[41-42,44-45] | 24 gpu | 1-00:00:00 | 6 | 2000 | 4 | Public |
| gpu_vbd | gpu_vbd | wn[46,49,51] | 14 gpu | 14-00:00:00 | 6 | 2000 | 2 | Private |
| gpu_hylab | gpu_hylab | wn44 | 4 gpu | 3-00:00:00 | 4 | 2000 | 1 | Private |
| gpu_fisstat | gpu_fisstat | wn50 | 4 gpu | 7-00:00:00 | 4 | 2000 | 1 | Private |
| gpu_guest | gpu_guest | wn[44,46] | 8 gpu | 1-00:00:00 | 6 | 200 | 2 | Guest |
| vrt | vrt | wn[61-64] | 1 core | 10-00:00:00 | 24 | 2000 | 1 | Public |
| mngt | all | UNLIMITED | Admin |
More details:
scontrol show partition
Private area : Quota Management - PBSpro - Slurm
Useful commands
Slurm commands:
scontrol show partition # Display the status of the partitions scontrol show job <jobid> # Display the status of a individual job sinfo # Groups of nodes and partitions sinfo -all # Detailed list of nodes groups sinfo -all | grep idle # List free nodes sinfo -N # List of nodes and their status srun <options> # Interactive mode sbatch <options> script.sh # Batch mode squeue # Display jobs in the queue squeue -all # Display jobs details in the queue sprio # View the factors that comprise a job's scheduling priority
For detailed information, refer to the man pages:
man sinfo man squeue
Custom commands:
hpc-sinfo hpc-sinfo-cpu hpc-sinfo-gpu hpc-show-qos hpc-show-user-account hpc-squeue
View information about Slurm nodes and partitions:
hpc-sinfo-cpu --partition=cpu hpc-sinfo-cpu --partition=cpu_guest hpc-sinfo-gpu --partition=gpu hpc-sinfo-gpu --partition=gpu_guest
View information about jobs located in the Slurm scheduling queue:
hpc-squeue --user=$USER hpc-squeue --user=<another_user> hpc-squeue --partition=cpu hpc-squeue --partition=cpu_guest hpc-squeue --partition=gpu hpc-squeue --partition=gpu_guest hpc-squeue --status=RUNNING hpc-squeue --states=PENDING
Main SLURM options
Partition
-p --partition=<partition name> Default: cpu Example: --partition=gpu
Qos
--qos=<qos name> Default: Same as the partition Example: --qos=gpu
Job Name
-J, --job-name=<jobname> Default: the name of the batch script
Nodes
-N, --nodes=<number of nodes> Example: --nodes=2 Do not share nodes with other running jobs: --exclusive
CPUs
-n, --ntasks=<global number of tasks> Example: -N2 -n4 # 4cpus on 2 nodes -c --cpus-per-task=<cpu per task> Example: -N2 -c2 # 2 nodes, each with 2 cpus --ntasks-per-node=<number of tasks per node> Example: --ntasks-per-node=8 ## 8 MPI tasks per node
GPUs
You can use one or more GPU by specifying a list of consumables:
--gres=[name:type:count] Example: --gres=gpu:p100:2 Example: --gres=gpu:a100_80g:2
If you don't specify the type of GPU, your job will run on p100:
--gres=[name:count] Example: --gres=gpu:2
Memory limit
Memory required per allocated CPU
--mem-per-cpu=<size{units}>
Example: --mem-per-cpu=1G
Default: 512M
Memory required per node (as an alternative to --mem-per-cpu)
--mem=<size{units}>
Example: --mem=4G
Time limit
Maximum execution time of the job. -t, --time=<days-hours:minutes:seconds> Default: --time=0-01:00:00 Example: --time=1-00:00:00
Account
Specifies the account to be charged for the used resources -A --account=<account> Default: see the command hpc-show-user-account Example: --account=T_HPC18A
See also TeamWork
Output and error
Output and error file name -o, --output=<filename pattern> -e, --error=<filename pattern> Example: --output=%x.o%j --error=%x.e%j ### %x=job name, %j=jobid Default: stdout and stderr are written in a file named slurm-<jobid>.out
One or more e-mail addresses, separated by commas, that will receive the notifications from the queue manager. --mail-user=<mail address> Default: University e-mail address Events generating the notification. --mail-type=<FAIL, BEGIN, END, TIME_LIMIT_50, TIME_LIMIT_90, NONE, ALL> Default: NONE Example: --mail-user=john.smith@unipr.it --mail-type=BEGIN,END
Export variables
Export new variable to the batch job. --export=<environment variables> Example: --export threads="8"
Priority
The priority (from queue to execution) is dynamically defined by three parameters:
- Timelimit
- Aging (waiting time in partition)
- Fair share (amount of resources used in last 14 days per account)
Advanced Resource Reservation
It is possible to define an advanced reservation for teaching activities or special requests.
To view active reservations:
scontrol show reservation
To use a reservation:
sbatch --reservation=<reservation name> nomescript.sh ## command line
or
#SBATCH --reservation=<reservation name> ## inside the script
Interactive example:
srun --reservation=corso_hpc18a --account=T_HPC18A -N1 -n1 -p vrt --pty bash ... exit
Batch example:
sbatch --reservation=corso_hpc18a --account=T_HPC18A -N1 -n2 nomescript.sh
In order to request a reservation send an e-mail to sti.calcolo@unipr.it.
Reporting/Accounting
Reporting
Synopsis:
hpc-report [--user=<user>] [--partition=<partition>] [--accounts=<accounts>] [--starttime=<start_time>] [--endtime=<end_time>]
hpc-report --help
Usage:
hpc-report
hpc-report --starttime=<start_time> --endtime=<end_time>
Example:
hpc-report --starttime=2022-10-19 --endtime=2022-11-02
hpc-report --starttime=2022-10-19 --endtime=2022-11-02 --user=$USER
Options:
-M, --cluster=<cluster> Filter results per cluster
-u, --user=<user> Filter results per user
-r, --partition=<partition> Filter results per partition
-A, --accounts=<accounts> Filter results per accounts
-S, --starttime=<start_time> Filter results per start date
-E, --endtime=<end_time> Filter results per end date
-N, --nodelist=<node_list> Filter results per node(s)
--csv-output=<csv> Save reports as CSV files
-h, --help Display a help message and quit
To see the list of user jobs executed in the last three days:
hpc-lastjobs
Job efficiency report:
seff <SLURM_JOB_ID>
Accounting
ToDo
Interactive jobs
Template command
Template command to run on the login node:
srun \
--partition=<partition> \
--qos=<qos> \
--nodes=<nodes> \
--ntasks-per-node=<ntasks_per_node> \
--account=<account> \
--time=<time> \
<other srun options> \
--pty \
bash
Interactive work session on the computing node:
#... some interactive activity on the node ... exit
Examples
Reserve 4 cores on 2 node on cpu partition:
srun --nodes=2 --ntasks-per-node=4 --partition=cpu --qos=cpu --pty bash
When the job starts you can run these commands:
echo "SLURM_JOB_NODELIS : $SLURM_JOB_NODELIST" # assigned nodes list echo "SLURM_NTASKS_PER_NODE: $SLURM_NTASKS_PER_NODE" # number of task per node scontrol show job $SLURM_JOB_ID # job details exit
Reserve 1 core on node wn34:
Implicit defaults: partition=cpu, qos=cpu, ntasks-per-node=1, timelimit=01:00:00, account=user's default, mem=512MB
srun --nodes=1 --nodelist=wn34 --pty bash
Reserve 1 chunk with 1 GPU p100 on gpu partition:
srun --nodes=1 --partition=gpu --qos=gpu --gres=gpu:p100:1 --mem=4G --pty bash
When the job starts you can run these commands:
echo "CUDA_VISIBLE_DEVICES: $CUDA_VISIBLE_DEVICES" exit
Reserve 2 cores on different nodes:
srun --nodes=2 --ntasks-per-node=1 --pty bash
Reserve 2 cores on the same node:
srun --nodes=1 --ntasks-per-node=2 --pty bash
Batch job
The commands to be executed on the node must be included in a shell script. To submit the job:
sbatch scriptname.sh
Slurm options can be specified in the command line or in the first lines of the shell script (see examples below).
Each job is assigned a unique numeric identifier <Job Id>.
At the end of the execution a file containing stdout and stderr will be created in the directory from which the job was submitted.
By default the file in named slurm-<job id>.out
If your job specify different partitions, scheduler assumes just the first one
Serial jobs on VRT
OpenMP example program: calculation of the product of two matrices
cp /hpc/share/samples/serial/mm.cpp .
- mm.cpp
/****************************************************************************** * FILE: mm.cpp * DESCRIPTION: * Serial Matrix Multiply - C Version * To make this a parallel processing progam this program would be divided into * two parts - the master and the worker section. The master task would * distributes a matrix multiply operation to numtasks-1 worker tasks. * AUTHOR: Blaise Barney * LAST REVISED: 04/02/05 * * g++ mm.cpp -o mm * ******************************************************************************/ #include <stdlib.h> #include <unistd.h> //optarg #include <time.h> //clock() #include <sys/time.h> // gettimeofday() #include <iostream> using namespace std; void options(int argc, char * argv[]); void usage(char * argv[]); int n; // dimensione lato matrice quadrata int main(int argc, char *argv[]) { n=1000; options(argc, argv); /* optarg management */ int NCB, NRA, NCA; NCB=n; NRA=n; NCA=n; int i, j, k; /* misc */ double *a, *b, *c; cout << "#Malloc matrices a,b,c " << n << "x" <<n << " of double \n"; a=(double *)malloc(sizeof(double)*NRA*NCA); b=(double *)malloc(sizeof(double)*NCA*NCB); c=(double *)malloc(sizeof(double)*NRA*NCB); clock_t t0, t1; double dt; struct timeval tempo; double tv1, tv2; cout << "#Start timer \n"; gettimeofday(&tempo,0); tv1=tempo.tv_sec+(tempo.tv_usec/1000000.0); t0 = clock(); cout << "#Initialize A, B, and C matrices \n"; for (i=0; i<NRA; i++) for (j=0; j<NCA; j++) *(a+i*NCA+j)= i+j; for (i=0; i<NCA; i++) for (j=0; j<NCB; j++) *(b+i*NCA+j)= i*j; for(i=0;i<NRA;i++) for(j=0;j<NCB;j++) *(c+i*NCA+j)= 0.0; cout << "#Perform matrix multiply\n"; for(i=0;i<NRA;i++) for(j=0;j<NCB;j++) for(k=0;k<NCA;k++) *(c+i*NCB+j)+= *(a+i*NCB+k) * *(b+k+j*NRA); t1 = clock();//time(NULL); gettimeofday(&tempo,0); tv2=tempo.tv_sec+(tempo.tv_usec/1000000.0); cout << "#Stop timer \n"; dt=(double)(t1-t0)/CLOCKS_PER_SEC; cout << "#Dim Time(clock) Time(elapled)\n" ; cout << n << " " << dt << " " << tv2-tv1 << endl; return 0; } /************************************************/ void options(int argc, char * argv[]) { int i; while ( (i = getopt(argc, argv, "n:h")) != -1) { switch (i) { case 'n': n = strtol(optarg, NULL, 10); break; case 'h': usage(argv); exit(1); case '?': usage(argv); exit(1); default: usage(argv); exit(1); } } } /***************************************/ void usage(char * argv[]) { cout << argv[0] << " [-n <dim_lato_matrice> ] [-h] " << endl; return; }
Script mm.bash for the submission stage:
- mm.bash
#!/bin/bash #SBATCH --output=%x.o%j ##SBATCH --error=%x.e%j #If error is not specified stderr is redirected to stdout #SBATCH --partition=vrt #SBATCH --qos=vrt #SBATCH --nodes=1 #SBATCH --cpus-per-task=1 ## Uncomment the following line if you need an amount of memory other than default (512MB) ##SBATCH --mem=2G ## Uncomment the following line if your job needs a wall clock time other than default (1 hour) ## Please note that priority of queued job decreases as requested time increases #SBATCH --time=0-00:30:00 ## Uncomment the following line if you want to use an account other than your default account ( see hpc-show-user-account ) ##SBATCH --account=<account> ## Print the list of the assigned resources echo "#SLURM_JOB_NODELIST: $SLURM_JOB_NODELIST" # default gcc version is 4.8.5. Load GNU module if you want to use a newer version of the GNU compiler # module load gnu g++ mm.cpp -o mm ./mm # Uncomment the following lines to compile and run using the INTEL compiler #module load intel #icc mm.cpp -o mm_intel #./mm_intel # Uncomment the following lines to compile and run using the PGI compiler #module load pgi #pgcc mm.cpp -o mm_pgi #./mm_pgi
Submission
sbatch mm.bash
Monitor the job status:
hpc-squeue (or squeue)
To cancel the job:
scancel <SLURM_JOB_ID>
Other serial examples
ls /hpc/share/samples/serial
OpenMP jobs on CPU
OpenMP example program:
cp /hpc/share/samples/omp/omp_hello.c .
- omp_hello.c
/****************************************************************************** * FILE: omp_hello.c * DESCRIPTION: * OpenMP Example - Hello World - C/C++ Version * In this simple example, the master thread forks a parallel region. * All threads in the team obtain their unique thread number and print it. * The master thread only prints the total number of threads. Two OpenMP * library routines are used to obtain the number of threads and each * thread's number. * AUTHOR: Blaise Barney 5/99 * LAST REVISED: 04/06/05 ******************************************************************************/ #include <omp.h> #include <stdio.h> #include <stdlib.h> int main (int argc, char *argv[]) { int nthreads, tid; /* Fork a team of threads giving them their own copies of variables */ #pragma omp parallel private(nthreads, tid) { /* Obtain thread number */ tid = omp_get_thread_num(); printf("Hello World from thread = %d\n", tid); /* Only master thread does this */ if (tid == 0) { nthreads = omp_get_num_threads(); printf("Number of threads = %d\n", nthreads); } } /* All threads join master thread and disband */ }
Script omp_hello.bash with the request for 28 CPUs:
- omp_hello.bash
#!/bin/bash #SBATCH --partition=cpu #SBATCH --qos=cpu #SBATCH --job-name=omp_hello-GNU_compiler #SBATCH --output=%x.o%j ##SBATCH --error=%x.e%j #If error is not specified stderr is redirected to stdout #SBATCH --nodes=1 #SBATCH --cpus-per-task=28 ## Uncomment the following line if you need an amount of memory other than default (512MB) ##SBATCH --mem=2G ## Uncomment the following line if your job needs a wall clock time other than default (1 hour) ## Please note that priority of queued job decreases as requested time increases ##SBATCH --time=0-00:30:00 ## Uncomment the following line if you want to use an account other than your default account (see hpc-show-user-account) ##SBATCH --account=<account> ##SBATCH --exclusive # uncomment to require a whole node with at least 28 cores echo "#SLURM_JOB_NODELIST: $SLURM_JOB_NODELIST" # Comment out the following line in case of exclusive request export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK echo "#OMP_NUM_THREADS : $OMP_NUM_THREADS" # Compile and run with the GNU compiler. Default gcc version is 4.8.5. # Load the gnu module if you want to use a newer version of the GNU compiler. #module load gnu gcc -fopenmp omp_hello.c -o omp_hello ./omp_hello # Uncomment the following lines to compile and run with the INTEL compiler #module load intel #icpc -qopenmp omp_hello.c -o omp_hello_intel #./omp_hello_intel # Uncomment the following lines to compile and run with the PGI compiler #module load pgi #pgc++ -mp omp_hello.c -o omp_hello_pgi #/omp_hello_pgi
Submission
sbatch omp_hello.bash
Other OpenMP examples
ls /hpc/share/samples/omp
MPI Jobs on CPU
MPI example program:
cp /hpc/share/samples/mpi/mpi_hello.c .
- mpi_hello.c
/****************************************************************************** * FILE: mpi_hello.c * DESCRIPTION: * MPI tutorial example code: Simple hello world program * AUTHOR: Blaise Barney * LAST REVISED: 03/05/10 ******************************************************************************/ #include "mpi.h" #include <stdio.h> #include <stdlib.h> #define MASTER 0 int main (int argc, char *argv[]) { int numtasks, taskid, len; char hostname[MPI_MAX_PROCESSOR_NAME]; MPI_Init(&argc, &argv); MPI_Comm_size(MPI_COMM_WORLD, &numtasks); MPI_Comm_rank(MPI_COMM_WORLD, &taskid); MPI_Get_processor_name(hostname, &len); printf ("Hello from task %d on %s!\n", taskid, hostname); if (taskid == MASTER) printf("MASTER: Number of MPI tasks is: %d\n", numtasks); MPI_Finalize(); }
Script mpi_hello.bash, 2 nodes with 4 CPUs per node:
- mpi_hello.bash
#!/bin/bash #SBATCH --partition=cpu #SBATCH --qos=cpu #SBATCH --job-name="mpi_hello-GNU_compiler" #SBATCH --output=%x.o%j ##SBATCH --error=%x.e%j #If error is not specified stderr is redirected to stdout #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 ## Uncomment the following line if you need an amount of memory other than default (512MB) ##SBATCH --mem=2G ## Uncomment the following line if your job needs a wall clock time other than default (1 hour) ## Please note that priority of queued job decreases as requested time increases ##SBATCH --time=0-00:30:00 ## Uncomment the following line if you want to use an account other than your default account ( see hpc-show-user-account ) ##SBATCH --account=<account> echo "# SLURM_JOB_NODELIST : $SLURM_JOB_NODELIST" echo "# SLURM_CPUS_PER_TASK : $SLURM_CPUS_PER_TASK" echo "# SLURM_JOB_CPUS_PER_NODE: $SLURM_JOB_CPUS_PER_NODE" # Gnu compiler module load gnu openmpi mpicc mpi_hello.c -o mpi_hello mpirun mpi_hello # Uncomment the following lines to compile and run using the INTEL compiler #module load intel intelmpi #mpicc mpi_mm.c -o mpi_mm_intel #mpirun mpi_hello_intel # Uncomment the following lines to compile and run using the PGI compiler #module load pgi/2018 openmpi/2.1.2/2018 #mpicc mpi_hello.c -o mpi_hello_pgi #mpirun --mca mpi_cuda_support 0 mpi_hello_pgi
Submission
sbatch mpi_hello.bash
Other MPI examples
ls /hpc/share/samples/mpi
MPI+OpenMP jobs on CPU
MPI + OpenMP example:
cp /hpc/share/samples/mpi+omp/mpiomp_hello.c .
- mpiomp_hello.c
#include <stdio.h> #include <mpi.h> #include <omp.h> int main(int argc, char *argv[]) { int numprocs, rank, namelen; char processor_name[MPI_MAX_PROCESSOR_NAME]; int iam = 0, np = 1; MPI_Init(&argc, &argv); MPI_Comm_size(MPI_COMM_WORLD, &numprocs); MPI_Comm_rank(MPI_COMM_WORLD, &rank); MPI_Get_processor_name(processor_name, &namelen); #pragma omp parallel default(shared) private(iam, np) { np = omp_get_num_threads(); iam = omp_get_thread_num(); printf("Hello from thread %3d out of %3d from process %3d out of %3d on %s\n", iam + 1, np, rank + 1, numprocs, processor_name); } MPI_Finalize(); }
Script mpiomp_hello.bash:
- mpiomp_hello.bash
#!/bin/sh #< 2 nodes, 2 MPI processes per node, 6 OpenMP threads per MPI process #SBATCH --partition=cpu #SBATCH --qos=cpu #SBATCH --job-name="mpi_hello-GNU_compiler" #SBATCH --output=%x.o%j ##SBATCH --error=%x.e%j #If error is not specified stderr is redirected to stdout #SBATCH --nodes=2 #SBATCH --ntasks-per-node=2 #< Number of MPI task per node #SBATCH --cpus-per-task=6 #< Number of OpenMP threads per MPI task ## Uncomment the following line if you need an amount of memory other than default (512MB) ##SBATCH --mem=2G ## Uncomment the following line if your job needs a wall clock time other than default (1 hour) ##SBATCH --time=0-00:30:00 ## Please note that priority of queued job decreases as requested time increases ## Uncomment the following line if you want to use an account other than your default account ##SBATCH --account=<account> echo "# SLURM_JOB_NODELIST : $SLURM_JOB_NODELIST" echo "# SLURM_CPUS_PER_TASK: $SLURM_CPUS_PER_TASK" export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK module load gnu openmpi mpicc -fopenmp mpiomp_hello.c -o mpiomp_hello mpirun mpiomp_hello ## Uncomment the following lines to compile and run with the INTEL compiler #module load intel impi #mpicc -qopenmp mpiomp_hello.c -o mpiomp_hello_intel #mpirun mpiomp_hello_intel
MPI for Python
Script hello-mpi-world.py:
- hello-mpi-world.py
#!/usr/bin/env python from mpi4py import MPI comm = MPI.COMM_WORLD name=MPI.Get_processor_name() print "Hello! I'm rank ",comm.rank," on node ", name, " from ", comm.size, " running in total" comm.Barrier() # wait for everybody to synchronize _here_
Submission example hello-mpi-world.sh:
- hello-mpi-world.sh
#!/bin/bash #SBATCH --partition=cpu #SBATCH --qos=cpu #SBATCH --output=%x.o%j #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 #SBATCH --account=T_HPC18A module load gnu7 openmpi3 py2-mpi4py #module load intel openmpi3 py2-mpi4py mpirun python -W ignore hello-mpi-world.py
Matrix-Vector product:
cp /hpc/group/T_HPC18A/samples/mpi4py/matrix-vector-product.py . cp /hpc/group/T_HPC18A/samples/mpi4py/matrix-vector-product.sh . sbatch matrix-vector-product.sh
GPU jobs
The gpu partition consists of:
- 2 machines with 6 gpu:p100 each
- 1 machine with 8 gpu:a100_80g
- 1 machine with 4 gpu:a100_40g
The GPUs of a single machine are identified by an integer ID that ranges from 0 to (N-1).
The compiler to use is nvcc:
Compilation example:
cp /hpc/share/samples/cuda/hello_cuda.cu . module load cuda nvcc hello_cuda.cu -o hello_cuda
- hello_cuda.cu
// This is the REAL "hello world" for CUDA! // It takes the string "Hello ", prints it, then passes it to CUDA with an array // of offsets. Then the offsets are added in parallel to produce the string "World!" // By Ingemar Ragnemalm 2010 #include <stdio.h> const int N = 7; const int blocksize = 7; __global__ void hello(char *a, int *b) { a[threadIdx.x] += b[threadIdx.x]; } int main() { char a[N] = "Hello "; int b[N] = {15, 10, 6, 0, -11, 1, 0}; char *ad; int *bd; const int csize = N*sizeof(char); const int isize = N*sizeof(int); printf("%s", a); cudaMalloc( (void**)&ad, csize ); cudaMalloc( (void**)&bd, isize ); cudaMemcpy( ad, a, csize, cudaMemcpyHostToDevice ); cudaMemcpy( bd, b, isize, cudaMemcpyHostToDevice ); dim3 dimBlock( blocksize, 1 ); dim3 dimGrid( 1, 1 ); hello<<<dimGrid, dimBlock>>>(ad, bd); cudaMemcpy( a, ad, csize, cudaMemcpyDeviceToHost ); cudaFree( ad ); printf("%s\n", a); return EXIT_SUCCESS; }
The gpu partition selection is done by specifying –partition=gpu and the selection of the gpu's type is done by specifying –gres=gpu:p100:<1-6> among the required resources.
Example of submission on 1 of the 6 GPUs p100 available on a single node of the gpu partition:
#!/bin/sh #< 1 node with 1 GPU #SBATCH --partition=gpu #SBATCH --qos=gpu #SBATCH --nodes=1 #SBATCH --gres=gpu:p100:1 #SBATCH --time=0-00:30:00 #SBATCH --mem=4G #< Charge resources to account ##SBATCH --account=<account> module load cuda echo "# SLURM_JOB_NODELIST : $SLURM_JOB_NODELIST" echo "# CUDA_VISIBLE_DEVICES: $CUDA_VISIBLE_DEVICES" ./hello_cuda
Example of submission on 1 of the 8 GPUs a100_80g available on a single node of the gpu partition:
#!/bin/sh #< 1 node with 1 GPU #SBATCH --partition=gpu #SBATCH --qos=gpu #SBATCH --nodes=1 #SBATCH --gres=gpu:a100_80g:1 #SBATCH --time=0-00:30:00 #SBATCH --mem=4G #< Charge resources to account ##SBATCH --account=<account> module load cuda echo "# SLURM_JOB_NODELIST : $SLURM_JOB_NODELIST" echo "# CUDA_VISIBLE_DEVICES: $CUDA_VISIBLE_DEVICES" ./hello_cuda
Example of submission of the N-BODY benchmark on all 6 GPUs p100 available in a single node of the gpu partition:
#!/bin/sh #< 1 node with 6 GPU #SBATCH --partition=gpu #SBATCH --qos=gpu #SBATCH --nodes=1 #SBATCH --gres=gpu:p100:6 #SBATCH --time=0-00:30:00 #SBATCH --mem=3G #< Charge resources to account ##SBATCH --account=<account> module load cuda echo "# SLURM_JOB_NODELIST : $SLURM_JOB_NODELIST" echo "# CUDA_VISIBLE_DEVICES: $CUDA_VISIBLE_DEVICES" /hpc/share/samples/cuda/nbody/nbody -benchmark -numbodies=1024000 -numdevices=7
In the case of N-BODY, the number of GPUs to be used is specified using the -numdevices option (the specified value must not exceed the number of GPUs required with the ngpus option).
In general, the GPU IDs to be used are derived from the value of the CUDA_VISIBLE_DEVICES environment variable.
In the case of the last example we have:
CUDA_VISIBLE_DEVICES=0,1,2,3,4,5,6
Teamwork
To share activities with other member of your group you can enter the command
newgrp <group>
The newgrp command modifies the primary group, modifies the file mode creation mask and sets the environmental variables GROUP and ARCHIVE for the duration of the session.
To change to the /hpc/group/<group> directory, execute the command
cd "$GROUP"
To change to the /hpc/archive/<group> directory, execute the command
cd "$ARCHIVE"
In the batch mode make sure the script to submit begins with
#!/bin/bash --login
and specify the account to use with the directive
#SBATCH --account=<account>
To display your default account
hpc-show-user-account
Job as different group
How to submit a batch job as different group without running the newgrp command?
Make sure the script to submit begins with
#!/bin/bash --login
Script slurm-super-group.sh:
- slurm-super-group.sh
#!/bin/bash --login #SBATCH --job-name=sg #SBATCH --output=%x.o%j #SBATCH --error=%x.e%j #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --mem=512M #SBATCH --partition=vrt #SBATCH --qos=vrt #SBATCH --time=0-00:05:00 #SBATCH --account=<account> shopt -q login_shell || exit 1 echo "user : $(id -un)" echo "group : $(id -gn)" echo "pwd : $(pwd)" test -n "$GROUP" && echo "GROUP : $GROUP" test -n "$ARCHIVE" && echo "ARCHIVE : $ARCHIVE" test -n "$SCRATCH" && echo "SCRATCH : $SCRATCH" test -n "$LOCAL_SCRATCH" && echo "LOCAL_SCRATCH: $LOCAL_SCRATCH"
Submitting slurm-super-group.sh:
sg <group> -c 'sbatch slurm-super-group.sh'
More information on the sg command:
man sg
Proxy
If you want to download files with http, https, ftp, git protocols make sure the script to submit begins with
#!/bin/bash --login
Script slurm-wget.sh:
- slurm-wget.sh
#!/bin/bash --login #SBATCH --job-name=wget #SBATCH --output=%x.o%j #SBATCH --error=%x.e%j #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --mem=512M #SBATCH --partition=vrt #SBATCH --qos=vrt #SBATCH --time=0-00:05:00 #SBATCH --account=<account> shopt -q login_shell || exit 1 echo $https_proxy wget -vc --no-check-certificate https://ftp.gnu.org/gnu/wget/wget-latest.tar.gz
Job Array
https://slurm.schedmd.com/job_array.html
Using a single SLURM script it is possible to submit a battery of Jobs, which can be executed in parallel, specifying a different numerical parameter for each submissive job.
The –array option specifies the numeric sequence of parameters. At each launch the value of the
parameter is contained in the $ SLURM_ARRAY_TASK_ID variable
Example:
Starts N job for the computing of Pi with a number of intervals increasing from 100000 to 900000 with increment of 10000:
cp /hpc/share/samples/serial/cpi/cpi_mc.c . gcc cpi_mc.c -o cpi_mc
Script slurm_launch_parallel.sh:
- slurm_launch_parallel.sh
#!/bin/sh #SBATCH --partition=vrt #SBATCH --qos=vrt #SBATCH --array=1-10:1 ## Charge resources to account ##SBATCH --account=<account> ##SBATCH --mem=4G N=$((${SLURM_ARRAY_TASK_ID}*1000000)) CMD="./cpi_mc -n $N" echo "# $CMD" eval $CMD
sbatch slurm_launch_parallel.sh
Gather the outputs:
grep -vh '^#' slurm_launch_parallel.sh.o*.*
Applications
Alphafold
Mathematica
Amber
Gromacs
Crystal
Schrodinger
Abaqus
Gaussian
Quantum-Espresso
XCrySDen
Q-Chem
IQmol
NAMD
ORCA
Echidna
Relion
CAVER
FDS
SRA Toolkit
Nextflow
CASTEP
GAMESS
Blender
Libraries
deal.II
Programming languages
MATLAB
Go
Julia
Python
R
Rust
Scala
Tools
Conda
Conda is an open source, cross-platform, language-agnostic package manager and environment management system.
Conda allows users to easily install different versions of binary software packages and any required libraries appropriate for their computing platform. Also, it allows users to switch between package versions and download and install updates from a software repository.
Conda is written in the Python programming language, but can manage projects containing code written in other languages (e.g., R), including multi-language projects. Conda can install the Python programming language, while similar Python-based cross-platform package managers (such as wheel or pip) cannot.
The HPC system includes predefined environments taylored for common problems, but users can build their own environments.
Containers
Singularity
Apptainer
Apptainer (formerly Singularity).
VTune
VTune is a performance profiler from Intel and is available on the HPC cluster.
General information from Intel: https://software.intel.com/en-us/get-started-with-vtune-linux-os
VTune local guide (work in progress)
RStudio
Rclone
Rclone is a command line program to manage files on cloud storage. It is a feature rich alternative to cloud vendors' web storage interfaces. Over 40 cloud storage products support rclone including S3 object stores, business and consumer file storage services, as well as standard transfer protocols.
OneDrive
Configuration for "Microsoft OneDrive"
ssh -X gui.hpc.unipr.it
As an alternative to ssx -X it is possible to open an Remote Desktop connection and use a terminal within the graphical desktop.
This activity only needs to be performed once:
module load rclone rclone config create onedrive onedrive
Help
rclone help rclone copy --help
Create directory, list, copy file
rclone mkdir onedrive:SharedWithSomeOne rclone ls onedrive:SharedWithSomeOne rclone copy <file> onedrive:SharedWithSomeOne
Share
Connect to Microsoft OneDrive and share the folder "SharedWithSomeOne".
Send the link to the people who will have access to the shared folder.
Optional: Mounting "onedrive" device on "OneDrive" folder
ssh login.hpc.unipr.it
module load rclone mkdir -p ~/OneDrive rclone mount onedrive: ~/OneDrive &
Unmounting "onedrive" device from "OneDrive" folder
fusermount -u -z ~/OneDrive
Share Point
Configuration for "Microsoft Sharepoint"
module load rclone rclone config
Follow the instructions:
- Press n for new remote
- Enter profile_name for rclone Sharepoint profile
- Enter 27 as Option Storage: 27 / Microsoft OneDrive \ "onedrive"
- Leave blank Client ID and OAuth Client Secret
- Enter 1 (Global) to Choose national cloud region for OneDrive.
- Enter no for "Edit Advanced Config"
- Enter yes for "Auto Config"
- Enter 3 as Config_type - Type of connection
/ Sharepoint site name or URL
3 | E.g. mysite or https://contoso.sharepoint.com/sites/mysite
\ "url"
- Enter https://univpr.sharepoint.com/sites/site_name as config_site_url
- Confirm the share point sites
Using profile_name is possible to copy file from or to share point Cloud Storage
rclone copy local_file_name profile_name:
rclone ls profile_name: